DMCCS Prediction
Log Out
This page provides access to CCS prediction models trained using a drug- and drug metabolite-specific CCS database (dmCCS). The models differ by the type of molecular descriptors (MDs) used during model traing, including 2D, 3D and combined feature sets. The 2D feature set, termed molecular quantum numbers (MQN), is a collection of MDs that capture compositional and topological information about a 2D chemical structure. The 3D feature set (MD3D) consists of principal moments of inertia and radial mass distribution computed from a 3D chemical structure modeled at a low level of theory (MMFF94). A specially selected combination of 2D and 3D features was used to train an additional prediction model (COMB). The CCS prediction models trained using these different feature sets differ in their performance characteristics due to the different information that is captured by the feature sets used to train them. The model(s) used for CCS prediction depends on which inputs are provided: SMILES structure is required for MQN and COMB models, and 3D structure is required for MD3D and COMB models. 3D structures should be computed using MMFF94 or similar level of theory in an ionized state (protonated or Na/K adduct) and uploaded in .mol2 format. SMILES structures should also be provided in an ionized state.
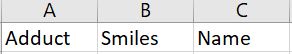
Make a CSV file containing information about your Predictions.
Then upload the CSV file below and click on "Make Predictions" to view the prediction results online
If you want to download the entire results in one file, please click “Download Results” directly
An example of the CSV file can be found below
**Make sure the header column names are as follows**